Building a custom CNN model: Identification of COVID-19
This article was published as a part of the Data Science Blogathon
Dear readers,
In this blog, let’s build our own custom CNN(Convolutional Neural Network) model all from scratch by training and testing it with our custom image dataset. This is, of course, mostly considered a more impressive work rather than training a pre-trained CNN model to classify images. You can learn about it if you are interested, from one of my previous blogs here. We will be using the validation-set approach to train the model and thus divide our dataset into training, validation, and testing datasets.
By the end of the blog, you will be able to build your own custom CNN model for COVID-19 to perform multi-class image classification by training it with your own dataset! Besides, we will also evaluate the trained model thoroughly on both validation and testing datasets by getting its classification report and confusion matrix. Moreover, we will also create a beautiful and simple front-end using Streamlit and integrate our model with the web application.
I have used Google Colab for all the implementation. Also, I have uploaded my dataset to my google drive for the project. The Streamlit web application can be easily launched from Google Colab itself.
So, let’s begin!
Agenda
- Introduction
- Applications
- Implementation
- Conclusion
Introduction
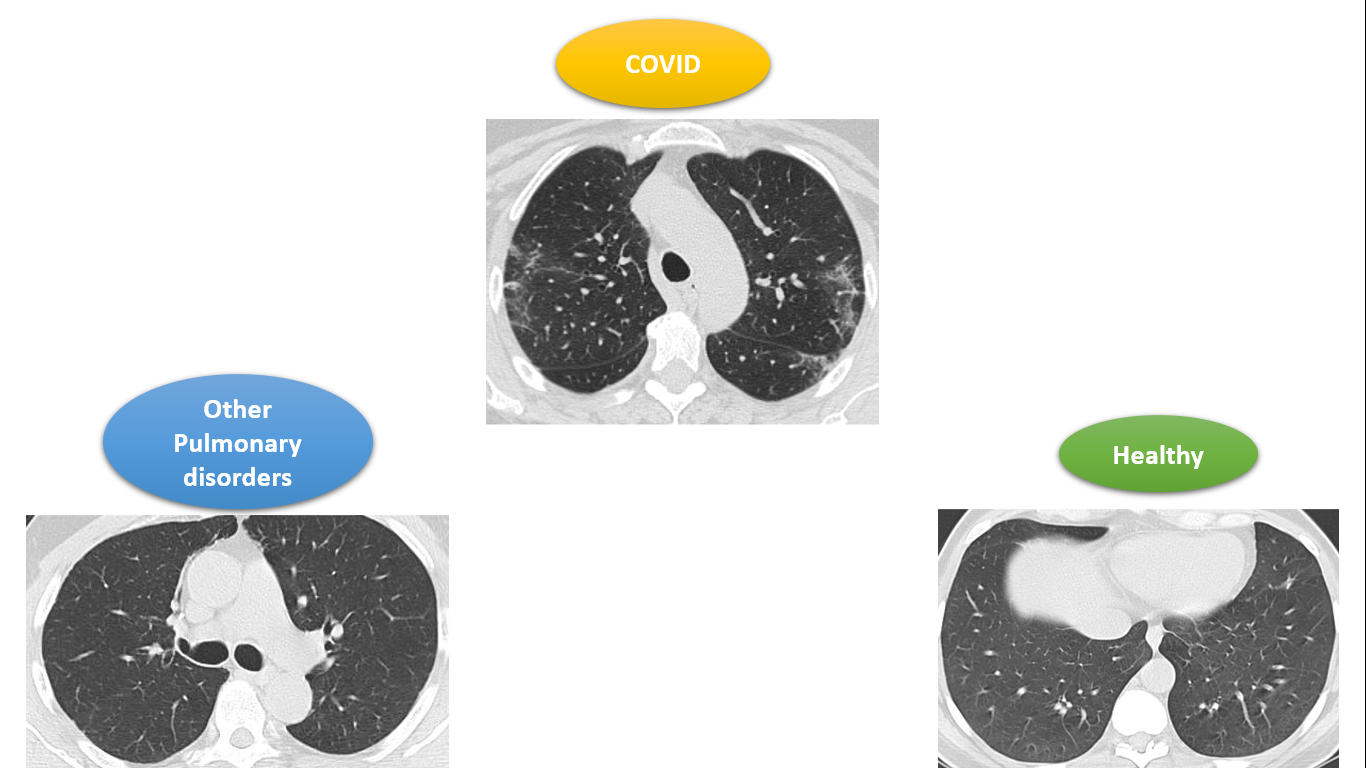
Firstly, a custom dataset is a dataset that is either prepared all by yourself like you are going out there in the city and clicking photographs to gather the images of your interest or downloading and using an open-source image dataset from renowned sites for datasets such as Kaggle, GitHub, etc. Summing up, it is your own dataset wherein you have stored images of required classes in the different folders-a folder for each class.
In this blog, I will be explaining how to build a CNN model for COVID-19 using TensorFlow on one of the COVID multiclass datasets of CT scans. It can be directly downloaded from here. Now, pause and ensure that you download the dataset to follow along with the implementation.
The given Kaggle dataset consists of chest CT scan images of patients suffering from the novel COVID-19, other pulmonary disorders, and those of healthy patients. For each of these three categories, there is a number of patients and for each of them, there is a number of CT scan images correspondingly.
We will be using these CT scan images to train our CNN model to identify if a given CT scan is that of a COVID patient, a patient suffering from other pulmonary disorders except COVID, or that of a healthy patient. The problem includes 3 classes namely: COVID, healthy, and other pulmonary disorders, shortly referred to as ‘others’.
Applications
We know that performing RTPCR to detect COVID is risky because the swab tests reach the throat via the nose which results in coughing and thus spreads virus particles into the air thereby putting the life of health workers at risk. Hence, CT scans are much safer than such swab tests, according to researchers. Besides, performing a CT scan test after the RTPCR test for a COVID-positive patient is recommended.
That’s where the project we are doing now can prove helpful for the medical community.
Implementation
Step-1: Image Pre-Processing
Step-2:Train-Test-Val split
Step-3: Model building
Step-4:Model evaluation
Step-5:Building the Streamlit Web Application
Firstly, let us import all the required packages as follows:
from tensorflow.keras.layers import Input, Lambda, Dense, Flatten,Dropout,Conv2D,MaxPooling2D from tensorflow.keras.models import Model from tensorflow.keras.preprocessing import image from sklearn.metrics import accuracy_score,classification_report,confusion_matrix from tensorflow.keras.preprocessing.image import ImageDataGenerator from sklearn.model_selection import train_test_split from tensorflow.keras.models import Sequential import numpy as np import pandas as pd import os import cv2 import matplotlib.pyplot as plt
Image Pre-Processing
Whenever we deal with image data, image pre-processing is the very first step and also the most crucial step to be performed. Here, we just reshape all the images to the desired size(100×100 in this project) and divide them by 255 as a step of normalization.
According to the directory structure of our dataset, as discussed in the previous section, we must go through every image present in folder-2(patient’s folder) which is further present in folder-1(the category folder-COVID, healthy, or others). Hence, the code for the same goes this way:
# re-size all the images to this
IMAGE_SIZE = (100,100)
path="/content/drive/MyDrive/MLH Project/dataset"
data=[]
c=0
for folder in os.listdir(path):
sub_path=path+"/"+folder
for folder2 in os.listdir(sub_path):
sub_path2=sub_path+"/"+folder2
for img in os.listdir(sub_path2):
image_path=sub_path2+"/"+img
img_arr=cv2.imread(image_path)
try:
img_arr=cv2.resize(img_arr,IMAGE_SIZE)
data.append(img_arr)
except:
c+=1
continue
print("Number of images skipped= ",c)
The below code performs normalization of the images:
x=np.array(data)
x=x/255.0
datagen = ImageDataGenerator(rescale = 1./255)
dataset = datagen.flow_from_directory(path,
target_size = IMAGE_SIZE,
batch_size = 32,
class_mode = 'sparse')
Further, to note the indices of the classes and assign these classes as labels, use the code below:
dataset.class_indices y=dataset.classes y.shape
Running the above code, you will observe that the following indices have been used for the corresponding classes:
|
|
|
|
|
|
|
|
NOTE: At the end of this step,all the images will be resized to 100×100 and although they are CT scans,they have been provided as color images in the Kaggle dataset chosen.So,thats why we get 100x100x3 when we try to see the shapes of x_train,x_val and x_test in the next section.Here,3 refers to the color image(R-G-B)
Train-Test-Val split
In this step, we will divide our dataset into a training set, testing set, and validation set in order to use the validation set approach to training our model to classify among the CT scans of COVID, healthy, or others.
We can use the traditional sklearn to achieve the same.
x_train,x_test,y_train,y_test=train_test_split(x,y,test_size=0.1) x_train,x_val,y_train,y_val=train_test_split(x_train,y_train,test_size=0.2)
Further, see the size of each of these datasets using the code below:
x_train.shape,y_train.shape x_val.shape,y_val.shape x_test.shape,y_test.shape
From the above code, you will observe that 3002 images belong to the train set, 751 images belong to the validation set and 418 images belong to the test set.
Step 3 Model Building
Now, we are all set to start coding our CNN model for COVID-19 from scratch. For this, we just need to keep adding layers, mostly Conv2D to extract features and MaxPooling2D to perform downsampling of the image. Besides, I have also used the BatchNormalization layer in order to improve the performance of the model in terms of its training as well as validation accuracies.
We can thus code our own CNN model as follows:
model=Sequential()
#covolution layer model.add(Conv2D(32,(3,3),activation='relu',input_shape=(100,100,3))) #pooling layer model.add(MaxPooling2D(2,2)) model.add(BatchNormalization())
#covolution layer model.add(Conv2D(32,(3,3),activation='relu')) #pooling layer model.add(MaxPooling2D(2,2)) model.add(BatchNormalization())
#covolution layer model.add(Conv2D(64,(3,3),activation='relu')) #pooling layer model.add(MaxPooling2D(2,2)) model.add(BatchNormalization())
#covolution layer model.add(Conv2D(64,(3,3),activation='relu')) #pooling layer model.add(MaxPooling2D(2,2)) model.add(BatchNormalization())
#i/p layer model.add(Flatten())
#o/p layer model.add(Dense(3,activation='softmax'))
model.summary()
A Convolutional Neural Network consists of several convolutional and pooling layers. I have added four Conv2D and MaxPooling layers. The first parameter of the Conv2D layer is where we must play a lot to arrive at the best possible model. You can read more about the syntax of Conv2D, MaxPooling2D, and BatchNormalization from the official Keras documentation.
After adding the convolution and max-pooling layers, I have included the BatchNormalization layers followed by which the input layer has been added using the Flatten() function.
There are no hidden layers here because they were not useful to improve the model’s performance during its training.
Finally, I have added the output layer which indeed gives us the output at the end! The Dense() function has been used for the same. It takes parameter 3 because we have 3 categories: COVID, healthy, and others. Also, the activation function used here is the softmax function because this is a multi-class problem.
This is just the model architecture. Now, before we train it, we must compile it as follows:
The optimizer used is the common adam optimizer. As the labels of the considered dataset are categorical and not one-hot-encoded, we must choose the sparse categorical cross-entropy loss function.
Early stopping is used to avoid overfitting. It stops training our model when it begins to overfit which in turn is identified through the sudden increase in the validation loss.
#compile model: model.compile(optimizer='adam',loss='sparse_categorical_crossentropy',metrics=['accuracy'])
The optimizer used is the widely used and the most preferred adam optimizer. As the labels of the considered dataset are categorical and not one-hot-encoded, we must choose the sparse categorical cross-entropy loss function.
Early stopping can be used to avoid overfitting. This is done as we don’t know how many epochs our model must be trained for.
from tensorflow.keras.callbacks import EarlyStopping early_stop=EarlyStopping(monitor='val_loss',mode='min',verbose=1,patience=5)
#Early stopping to avoid overfitting of model
Now, lets finally train our custom CNN model for, say,30 epochs:
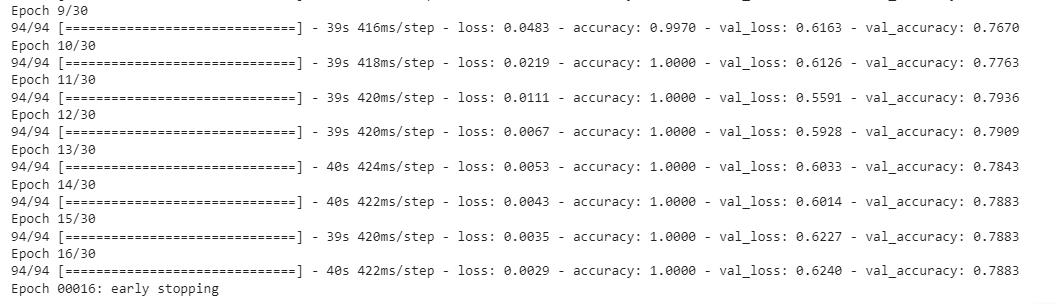
history=model.fit(x_train,y_train,validation_data=(x_val,y_val),epochs=30,callbacks=[early_stop],shuffle=True)
Early stopping was encountered at the 16th epoch itself and thus model was trained only for 16 epochs, at the end of which it showed a training accuracy of 100% and a validation accuracy of 78.83%.
Model Evaluation
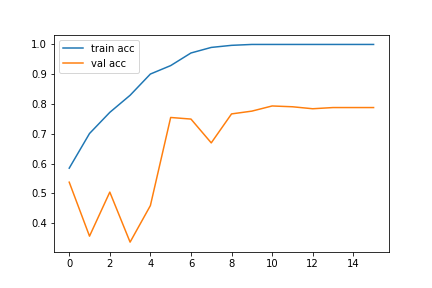
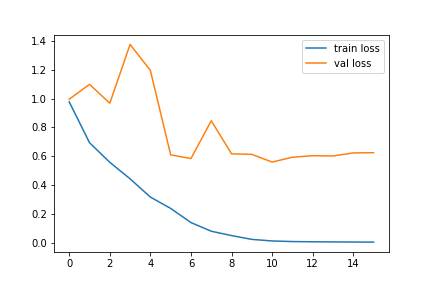
The best way to visualize our model training is by using the loss and accuracy graphs. The following codes can be used to get the loss and accuracy graphs for our trained model:
#loss graph plt.plot(history.history['loss'],label='train loss') plt.plot(history.history['val_loss'],label='val loss') plt.legend()
plt.savefig('loss-graph.png')
plt.show()
# accuracies plt.plot(history.history['accuracy'], label='train acc') plt.plot(history.history['val_accuracy'], label='val acc') plt.legend()
plt.savefig('acc-graph.png')
plt.show()
The accuracy and loss graphs in my case are as follows:
Classification report and confusion matrix for the validation dataset:
y_val_pred=model.predict(x_val) y_val_pred=np.argmax(y_val_pred,axis=1) print(classification_report(y_val_pred,y_val))
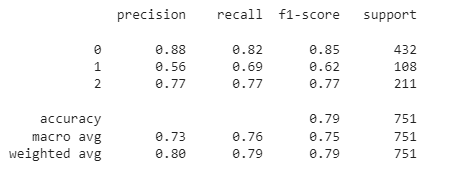
confusion_matrix(y_val_pred,y_val)

Therefore, it can be clearly concluded that our CNN model for COVID CT scans is the best. It shows an average performance for the class of other pulmonary disorders. However, its performance is comparatively poor for healthy patients. Besides, our model shows an accuracy of 79% on the validation dataset.
Classification report and confusion matrix for the test dataset, which is completely new to our model:
y_pred=model.predict(x_test) y_pred=np.argmax(y_pred,axis=1) print(classification_report(y_pred,y_test)) confusion_matrix(y_pred,y_test)
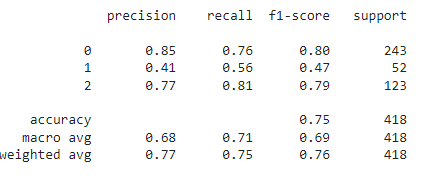

It shows an accuracy of 75% on the test dataset with a similar performance to that of the validation dataset.
Overall, we can conclude that we have developed a realistic CNN model for COVID-19 all from scratch.
Lets us now save the model with the following code:
model.save('/content/drive/MyDrive/MLH Project/model-recent.h5')
Building the Streamlit Web Application
In this step, we will create a front-end using Streamlit where the user can upload an image of a chest CT scan. Clicking the ‘Predict’ button pre-processes the input image to 100×100, which is the input shape for our CNN model for COVID-19, and then sends it to our model.
To check which category our model predicted this image to be, we get the index corresponding to the maximum value using the np.argmax() function and thus conclude according to the codes of the labels discussed in the table of step 1.
Firstly, we must install Streamlit and import ngrok:
!pip install streamlit --quiet !pip install pyngrok==4.1.1 --quiet from pyngrok import ngrok
Then comes the actual code! Here, we mainly load the saved model-the h5 file and predict using it. The name of the model file is model-recent.h5.There is an option to upload an image directly from your local system and check its category-if the CT scan is that of COVID or healthy or other pulmonary disorders.
st. button(‘Predict’) creates a button with “Predict” written in it and returns True when the user clicks the button.st.title() makes the text in its parameter, to be displayed in dark bold. These are some of the Streamlit functions to talk about.
%%writefile app.py import streamlit as st import tensorflow as tf
import numpy as np from PIL import Image # Strreamlit works with PIL library very easily for Images import cv2
model_path='/content/drive/MyDrive/MLH Project/model-recent.h5'
st.title("COVID-19 Identification Using CT Scan")
upload = st.file_uploader('Upload a CT scan image')
if upload is not None:
file_bytes = np.asarray(bytearray(upload.read()), dtype=np.uint8)
opencv_image = cv2.imdecode(file_bytes, 1)
opencv_image = cv2.cvtColor(opencv_image,cv2.COLOR_BGR2RGB) # Color from BGR to RGB
img = Image.open(upload)
st.image(img,caption='Uploaded Image',width=300)
if(st.button('Predict')):
model = tf.keras.models.load_model(model_path)
x = cv2.resize(opencv_image,(100,100))
x = np.expand_dims(x,axis=0)
y = model.predict(x)
ans=np.argmax(y,axis=1)
if(ans==0):
st.title('COVID')
elif(ans==1):
st.title('Healthy')
else:
st.title('Other Pulmonary Disorder')
Finally, get the URL of your web application from:
!nohup streamlit run app.py & url = ngrok.connect(port='8501') url
Paste this URL in the Chrome web browser to see our beautiful application.
Results
Your web application should look like this after you upload an image using the Browse button and then click the Predict button.
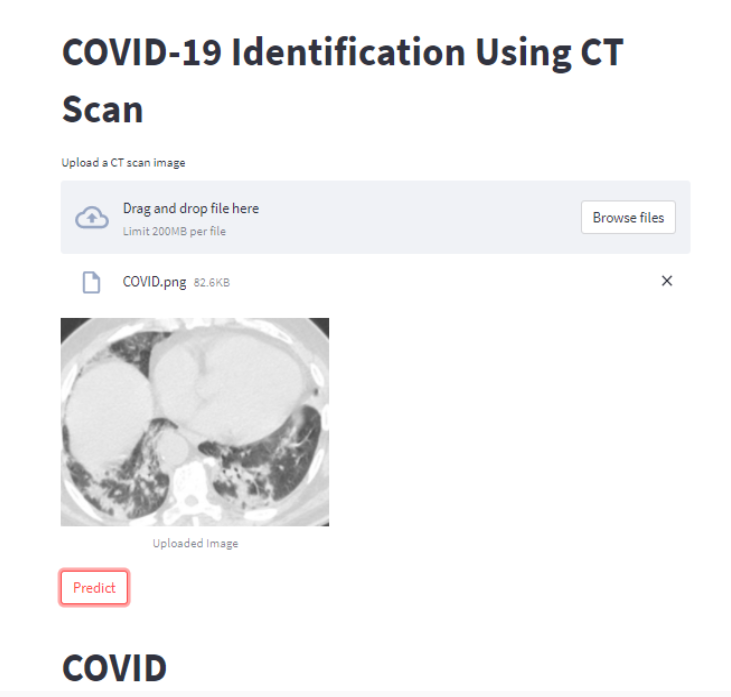
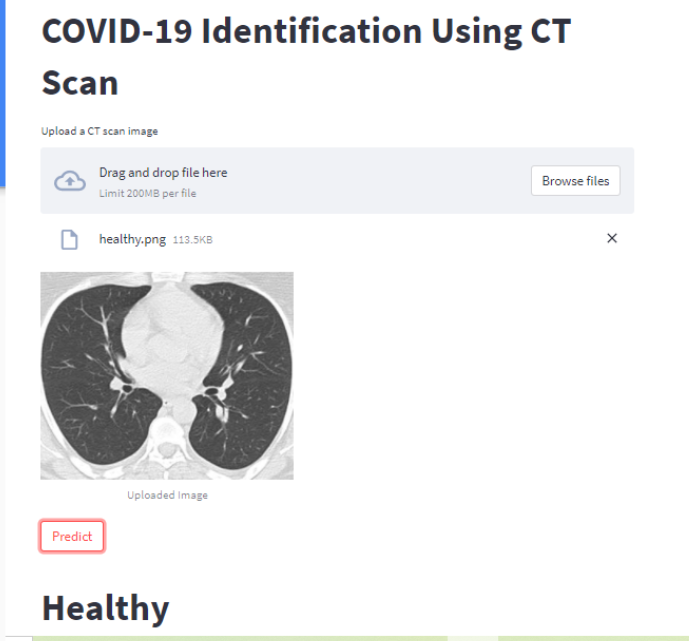
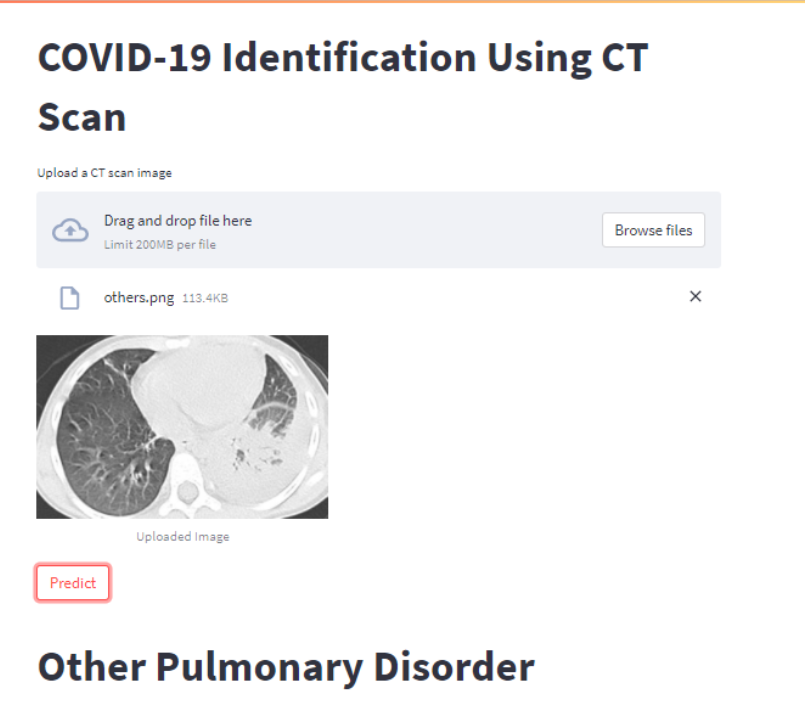
Conclusion
Thus, we successfully built and trained our own CNN model for COVID-19 with our dataset! The same approach can be used for two or more classes. All you have to do is change the number of classes in the output layer or the last layer of the model architecture.
That’s it!
Hope you liked my blog and found it useful!
You can get the entire code from here.
Thanks for reading!
References
About Me
I am Nithyashree V, a final year BTech Computer Science and Engineering student. I love learning such cool technologies and putting them into practice, especially observing how they help us solve society’s challenging problems. My areas of interest include Artificial Intelligence, Data Science, and Natural Language Processing.
Here is my LinkedIn profile: My LinkedIn
You can read my other articles on Analytics Vidhya from here.









Please share the dataset link.
Dear Nithyashree , Congrats on your article "Building a custom CNN model: Identification of COVID-19". It was very well written and easy to follow too. (and the VGG model article was also very good!!). I wanted to work with the Covid19 model for a class demonstration assignment. However, i could not as i did not get the link for the dataset you have used. You state in the article that the dataset can be downloaded from "here", but the url link does not exist in the article. I would greatly appreciate it if you could provide me the link to the Covid19 dataset used in the article. It would be very helpful, rather than create a new data set to fit the model.. Looking forward to hearing from you. Best Regards, Srikar
Hello Srikar. Thank you for your compliments. Here is the dataset link: https://www.kaggle.com/plameneduardo/a-covid-multiclass-dataset-of-ct-scans
Guys,here is the dataset link: https://www.kaggle.com/plameneduardo/a-covid-multiclass-dataset-of-ct-scans
Hello Dear Nithyashree, I have recently been working on deep learning and I have implemented your program on the Kagge and I encountered this error while running in the data sharing section. please guide me. ValueError Traceback (most recent call last) /tmp/ipykernel_36/4152827673.py in ----> 1 x_train,x_test,y_train,y_test=train_test_split(x,y,test_size=0.1) 2 x_train,x_val,y_train,y_val=train_test_split(x_train,y_train,test_size=0.2) /opt/conda/lib/python3.7/site-packages/sklearn/model_selection/_split.py in train_test_split(*arrays, **options) 2125 raise TypeError("Invalid parameters passed: %s" % str(options)) 2126 -> 2127 arrays = indexable(*arrays) 2128 2129 n_samples = _num_samples(arrays[0]) /opt/conda/lib/python3.7/site-packages/sklearn/utils/validation.py in indexable(*iterables) 290 """ 291 result = [_make_indexable(X) for X in iterables] --> 292 check_consistent_length(*result) 293 return result 294 /opt/conda/lib/python3.7/site-packages/sklearn/utils/validation.py in check_consistent_length(*arrays) 254 if len(uniques) > 1: 255 raise ValueError("Found input variables with inconsistent numbers of" --> 256 " samples: %r" % [int(l) for l in lengths]) 257 258 ValueError: Found input variables with inconsistent numbers of samples: [210, 4171] Thanks