Diminishing the Dimensions with PCA!
This article was published as a part of the Data Science Blogathon.
Introduction
This article will give you clarity on what is dimensionality reduction, its need, and how it works. There is also python code for illustration and comparison between PCA, ICA, and t-SNE.
What is Dimensionality Reduction and why do we need it?
Dimensionality Reduction is simply reducing the number of features (columns) while retaining maximum information. Following are reasons for Dimensionality Reduction:
- Dimensionality Reduction helps in data compression, and hence reduced storage space.
- It reduces computation time.
- It also helps remove redundant features, if any.
- Removes Correlated Features.
- Reducing the dimensions of data to 2D or 3D may allow us to plot and visualize it precisely. You can then observe patterns more clearly.
- It is helpful in noise removal also and as a result of that, we can improve the performance of models.
How does Dimensionality Reduction works (using PCA)?
There are many methods for Dimensionality Reduction like PCA, ICA, t-SNE, etc., we shall see PCA (Principal Component Analysis).
Let’s first understand what is information in data. Consider the following imaginary data, which has Age, Weight, and Height of people.
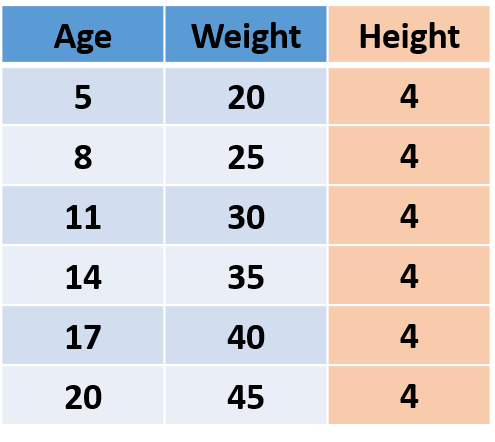
Fig.1
The information lies in the variance!
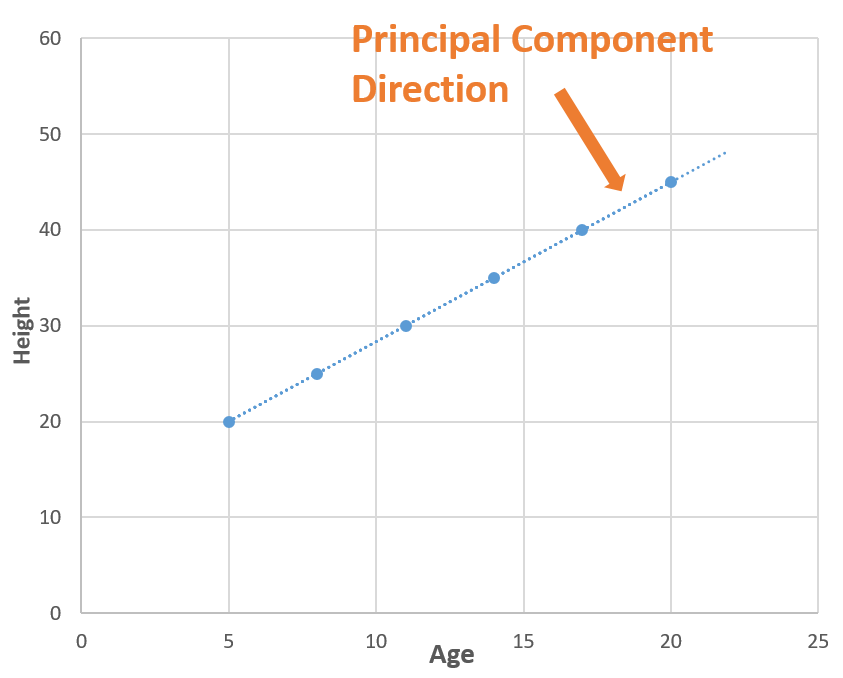
Now consider a line (blue dotted line) that is passing through all the points. The blue dotted line is capturing all the information so, we can replace ‘Age’ and ‘Weight’ (2 Dimensions) with the blue dotted line (1 Dimension) without losing any information, and in this way, we have done dimensionality reduction (2 dimensions to 1 dimension). The blue dotted line is called Principal Component.
You make be wondering how did we find the blue dotted line (Principal Component), so there is a lot of mathematics playing behind it. You can read this article which explains this, but in simple words, we find the directions in which we can capture maximum variance using Eigenvalues and Eigenvectors.
PCA illustration in Python
Let’s see how to use PCA with an example. I have used wine quality data which has 12 features like acidity, residual sugar, chlorides, etc., and the quality of the wine for 1600 samples. You can download the jupyter notebook from here and follow along.
First, let’s load the data.
import numpy as np import pandas as pd from sklearn.decomposition import PCA
df = pd.read_csv('winequality-red.csv')
df.head()
y = df.pop('quality')
After loading the data, we removed the ‘Quality’ of the wine as it is the target feature. Now we will scale the data because if not PCA will not be able to find the optimal Principal Components. For eg. we have a feature in ‘meters’ and another in ‘kilometers’, the feature with unit ‘meter’ will have more variance than ‘kilometers’ (1 km = 1000 m), so PCA will give more importance to the feature with high variance. So, let’s do the scaling.
from sklearn.preprocessing import StandardScaler scalar = StandardScaler() df_scaled = pd.DataFrame(scalar.fit_transform(df), columns=df.columns) df_scaled
Now we are ready to apply for PCA.
pca = PCA() df_pca = pd.DataFrame(pca.fit_transform(df_scaled)) df_pca
After applying PCA, we got 11 Principal Components. It’s interesting to see how much variance each principal component captures.
import matplotlib.pyplot as plt
pd.DataFrame(pca.explained_variance_ratio_).plot.bar()
plt.legend('')
plt.xlabel('Principal Components')
plt.ylabel('Explained Varience');
The first 5 Principal Components are capturing around 80% of the variance so we can replace the 11 original features (acidity, residual sugar, chlorides, etc.) with the new 5 features having 80% of the information. So, we have reduced the 11 dimensions to only 5 dimensions while retaining most of the information.
Difference between PCA, ICA, and t-SNE
PCA can also be used for clustering. I have used the iris dataset for this purpose. The following image will show you the clustering using PCA, ICA, and t-SNE.
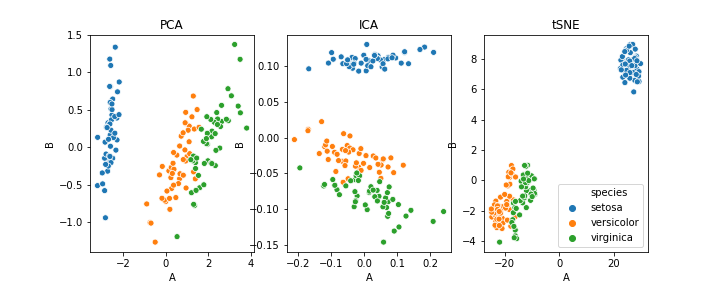
As this article focuses on the basic ideas behind dimensionality reduction and working of PCA, we will not go into the details but those who are interested can go through this article.
In short, PCA does dimensionality reduction while capturing most of the variance. Now, you would be excited to apply PCA to your data but before that, I want to highlight some of the disadvantages of PCA:
- Independent variables become less interpretable
- Data standardization is a must before PCA
- Information Loss
Hope you find this article informative. Thank you!
The media shown in this article are not owned by Analytics Vidhya and is used at the Author’s discretion.








