Introduction
Machine learning models aim to understand patterns within data, enabling predictions, answers to questions, or a deeper understanding of concealed patterns. This iterative learning process involves the model acquiring patterns, testing against new data, adjusting parameters, and repeating until achieving satisfactory performance. The evaluation phase, essential for regression problems, employs loss functions. As a data scientist, it’s crucial to monitor regression metrics like mean squared error and R-squared to ensure the model doesn’t overfit the training data. Libraries like scikit-learn provide tools to train and evaluate regression models, helping data scientists build effective solutions.
Learning Objectives:
- Understand the role of loss functions in evaluating regression models.
- Learn about the different types of regression loss functions and their applications.
- Identify the pros and cons of various regression evaluation metrics.
- Gain hands-on experience implementing regression metrics using Python libraries.
- Develop the ability to select appropriate loss functions based on specific data characteristics and modeling needs.
This article was published as a part of the Data Science Blogathon.
Table of contents
- Role of Loss Functions in Model Evaluation
- Crucial Factors in Choosing Loss Functions
- List of Top 13 Evaluation Metrics
- Mean Absolute Error (MAE)
- Mean Bias Error (MBE)
- Relative Absolute Error (RAE)
- Mean Absolute Percentage Error (MAPE)
- Mean Squared Error (MSE)
- Root Mean Squared Error (RMSE)
- Relative Squared Error (RSE)
- Normalized Root Mean Squared Error (NRMSE)
- Relative Root Mean Squared Error (RRMSE)
- Root Mean Squared Logarithmic Error (RMSLE)
- Huber Loss
- Log Cosh Loss
- Quantile Loss
- Frequently Asked Questions
Role of Loss Functions in Model Evaluation
Loss functions compare the model’s predicted values with actual values, gauging its efficacy in mapping the relationship between X (feature) and Y (target). The loss, indicating the disparity between predicted and actual values, guides model refinement. A higher loss denotes poorer performance, demanding adjustments for optimal training.
Crucial Factors in Choosing Loss Functions
Selecting an appropriate loss function hinges on various factors such as the algorithm, data outliers, and the need for differentiability. With many options available, each with distinct properties, there is no universal solution. This article comprehensively lists regression loss functions, outlining their advantages and drawbacks. Implementable across various libraries, the code examples use NumPy to enhance the underlying mechanisms’ transparency.
Let’s delve into the world of regression loss functions without delay.
List of Top 13 Evaluation Metrics
Here is a list of 13 evaluation metrics
- Mean Absolute Error (MAE)
- Mean Bias Error (MBE)
- Relative Absolute Error (RAE)
- Mean Absolute Percentage Error (MAPE)
- Mean Squared Error (MSE)
- Root Mean Squared Error (RMSE)
- Relative Squared Error (RSE)
- Normalized Root Mean Squared Error (NRMSE)
- Relative Root Mean Squared Error (RRMSE)
- Root Mean Squared Logarithmic Error (RMSLE)
- Hyber Loss
- Log Cosh Loss
- Quantile Loss
Mean Absolute Error (MAE)
Mean absolute error, or L1 loss, stands out as one of the simplest and easily comprehensible loss functions and evaluation metrics. It computes by averaging the absolute differences between predicted and actual values across the dataset. Mathematically, it represents the arithmetic mean of absolute errors, focusing solely on their magnitude, irrespective of direction. A lower MAE indicates superior model accuracy.
MAE formula is:
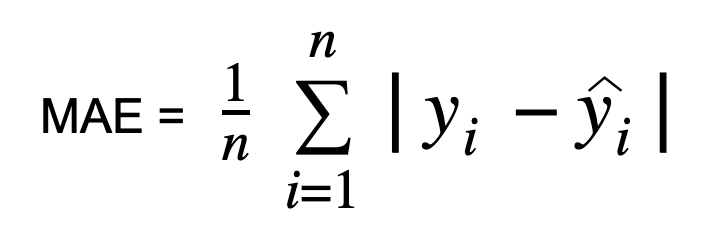
where
- y_i = actual value
- y_hat_i = predicted value
- n = sample size
Python Code:
import numpy as np
def mean_absolute_error(true, pred):
"""
Calculates the Mean Absolute Error (MAE) between the true and predicted values.
Args:
true (numpy.ndarray): An array of true values.
pred (numpy.ndarray): An array of predicted values.
Returns:
float: The Mean Absolute Error.
"""
mae = np.mean(np.abs(true - pred))
return mae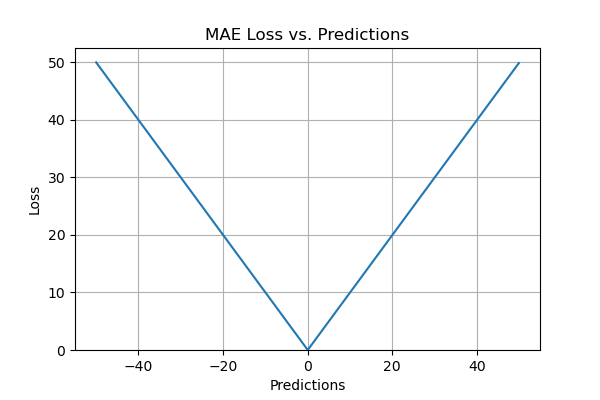
Pros of the MAE Evaluation Metric
- It is an easy-to-calculate evaluation metric.
- All the errors are weighted on the same scale since absolute values are taken.
- It is useful if the training data has outliers as MAE does not penalize high errors caused by outliers.
- It provides an even measure of how well the model is performing.
Cons of the MAE evaluation metric
- Sometimes the large errors coming from the outliers end up being treated as the same as low errors.
- MAE follows a scale-dependent accuracy measure using the same scale as the data being measured. Hence it cannot be used to compare series’ using different measures.
- One of the main disadvantages of MAE is that it is not differentiable at zero. Many optimization algorithms tend to use differentiation to find the optimum value for parameters in the evaluation metric.
- It can be challenging to compute gradients in MAE.
Mean Bias Error (MBE)
In “Mean Bias Error,” bias reflects the tendency of a measurement process to overestimate or underestimate a parameter. It has a single direction, positive or negative. Positive bias implies an overestimated error, while negative bias implies an underestimated error. Mean Bias Error (MBE) calculates the mean difference between predicted and actual values, quantifying overall bias without considering absolute values. Similar to MAE, MBE differs in not taking the absolute value. Caution is needed with MBE, as positive and negative errors can cancel each other out.
The formula for MBE:
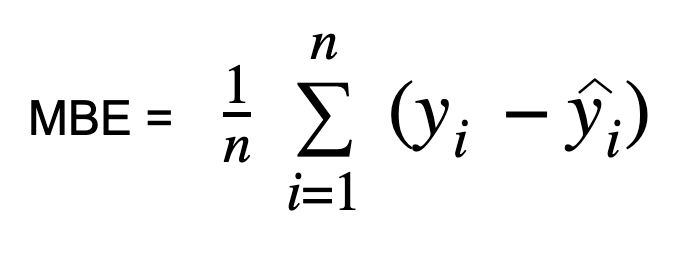
def mean_bias_error(true, pred):
bias_error = true - pred
mbe_loss = np.mean(np.sum(diff) / true.size)
return mbe_loss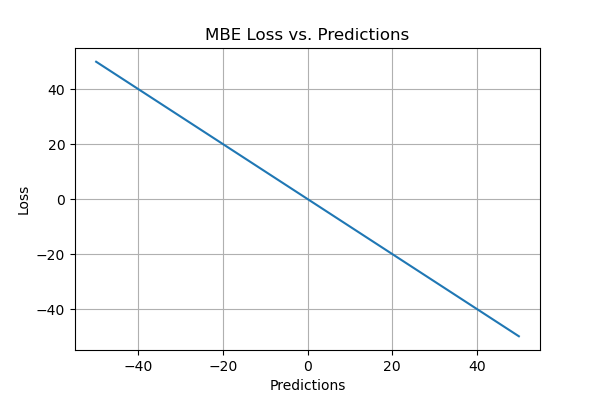
Pros of the MBE Evaluation Metric
- MBE is a good measure if you want to check the direction of the model (i.e. whether there is a positive or negative bias) and rectify the model bias.
Cons of the MBE Evaluation Metric
- It is not a good measure in terms of magnitude as the errors tend to compensate each other.
- It is not highly reliable because sometimes high individual errors produce low MBE.
- As an evaluation metric, it can be consistently wrong in one direction. For example, if you’re trying to predict traffic patterns it always shows lower traffic than what is observed.
Relative Absolute Error (RAE)
Relative root mean square error Absolute Error is calculated by dividing the total absolute error by the absolute difference between the mean and the actual value. The formula for RAE is:
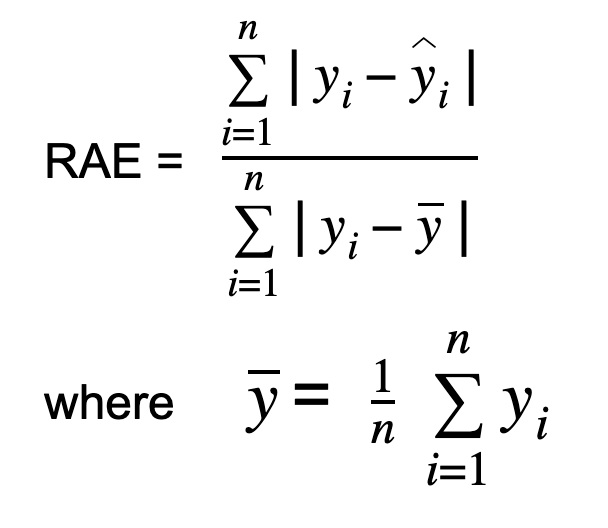
where y_bar is the mean of the n actual values.
RAE measures the performance of a predictive model and is expressed in terms of a ratio. The value of RAE can range from zero to one. A good model will have values close to zero, with zero being the best value. This error shows how the mean residual relates to the mean deviation of the target function from its mean.
def relative_absolute_error(true, pred):
true_mean = np.mean(true)
squared_error_num = np.sum(np.abs(true - pred))
squared_error_den = np.sum(np.abs(true - true_mean))
rae_loss = squared_error_num / squared_error_den
return rae_loss
Pros of the RAE Evaluation Metric
- RAE can be used to compare models where errors are measured in different units.
- In some cases, RAE is reliable as it offers protection from outliers.
Cons of the RAE Evaluation Metric
- One main drawback of RAE is that it can be undefined if the reference forecast is equal to the ground truth.
Mean Absolute Percentage Error (MAPE)
Calculate Mean Absolute Percentage Error (MAPE) by dividing the absolute difference between the actual and predicted values by the actual value. This absolute percentage is averaged across the dataset. MAPE, also known as Mean Absolute Percentage Deviation (MAPD), increases linearly with error. Lower MAPE values indicate better model performance.
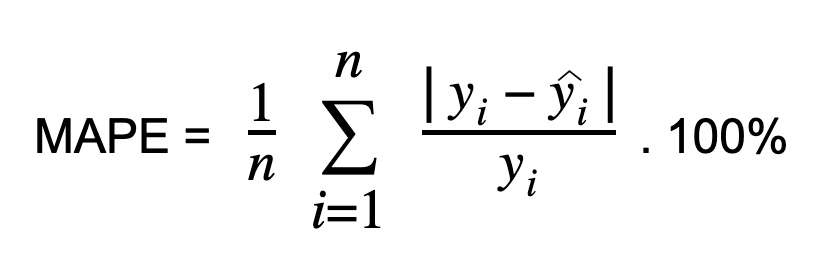
def mean_absolute_percentage_error(true, pred):
abs_error = (np.abs(true - pred)) / true
sum_abs_error = np.sum(abs_error)
mape_loss = (sum_abs_error / true.size) * 100
return mape_loss
Pros of the MAPE Evaluation Metric
- MAPE is independent of the scale of the variables since its error estimates are in terms of percentage.
- All errors are normalized on a common scale and it is easy to understand.
- As MAPE uses absolute percentage errors, the problem of positive values and negative values canceling each other out is avoided.
Cons of the MAPE Evaluation Metric
- MAPE faces a critical problem when the denominator becomes zero, resulting in a “division by zero” challenge.
- MAPE exhibits bias by penalizing negative errors more than positive errors, potentially favoring methods with lower values.
- Due to the division operation, MAPE’s sensitivity to alterations in actual values leads to varying losses for the same error. For example, an actual value of 100 and a predicted value of 75 results in a 25% loss, while an actual value of 50 and a predicted value of 75 yields a higher 50% loss, despite the identical error of 25.
Mean Squared Error (MSE)
MSE is one of the most common regression loss functions and an important error metric. In Mean Squared Error, also known as L2 loss, we calculate the error by squaring the difference between the predicted value and actual value and averaging it across the dataset.
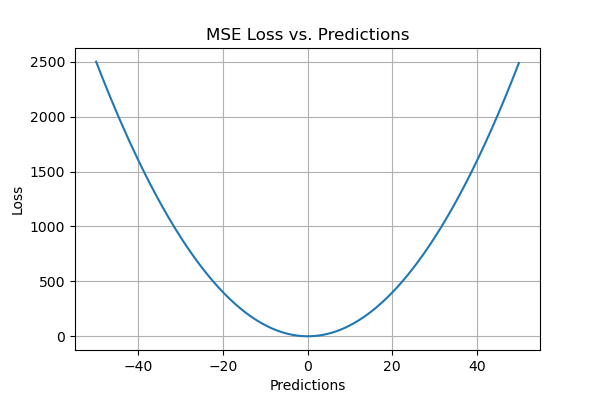
MSE is also known as Quadratic loss as the penalty is not proportional to the error but to the square of the error. Squaring the error gives higher weight to the outliers, which results in a smooth gradient for small errors.
Optimization algorithms benefit from this penalization for large errors as it helps find the optimum values for parameters using the least squares method. MSE will never be negative since the errors are squared. The value of the error ranges from zero to infinity. MSE increases exponentially with an increase in error. A good model will have an MSE value closer to zero, indicating a better goodness of fit to the data.
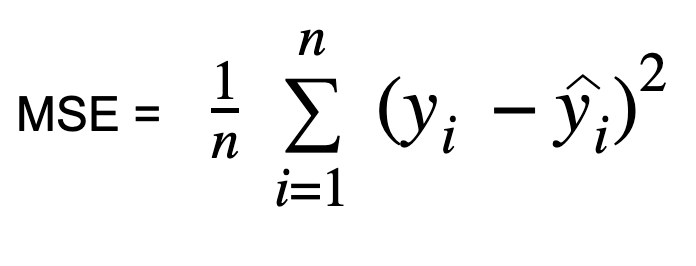
def mean_squared_error(true, pred):
squared_error = np.square(true - pred)
sum_squared_error = np.sum(squared_error)
mse_loss = sum_squared_error / true.size
return mse_loss
Pros of the MSE Evaluation Metric
- MSE values are expressed in quadratic equations. Hence when we plot it, we get a gradient descent with only one global minima.
- For small errors, it converges to the minima efficiently. There are no local minima.
- MSE penalizes the model for having huge errors by squaring them.
- It is particularly helpful in weeding out outliers with large errors from the model by putting more weight on them.
Cons of the MSE Evaluation Metric
- One of the advantages of MSE becomes a disadvantage when there is a bad prediction. The sensitivity to outliers magnifies the high errors by squaring them.
- MSE will have the same effect for a single large error as too many smaller errors. But mostly we will be looking for a model which performs well enough on an overall level.
- MSE is scale-dependent as its scale depends on the scale of the data. This makes it highly undesirable to compare different measures.
- When a new outlier is introduced into the data, the model will try to take in the outlier. By doing so it will produce a different line of best fit which may cause the final results to be skewed.
Root Mean Squared Error (RMSE)
Root Mean Square Error in Machine Learning (RMSE) is a popular metric used in machine learning and statistics to measure the accuracy of a predictive model. It quantifies the differences between predicted values and actual values, squaring the errors, taking the mean, and then finding the square root. RMSE provides a clear understanding of the model’s performance, with lower values indicating better predictive accuracy relative root mean square error.
It is computed by taking the square root of MSE. RMSE is also called the Root Mean Square Deviation. It measures the average magnitude of the errors and is concerned with the deviations from the actual value. RMSE value with zero indicates that the model has a perfect fit. The lower the RMSE, the better the model and its predictions. A higher relative root mean square error in machine learning indicates that there is a large deviation from the residual to the ground truth. RMSE can be used with different features as it helps in figuring out if the feature is improving the model’s prediction or not.
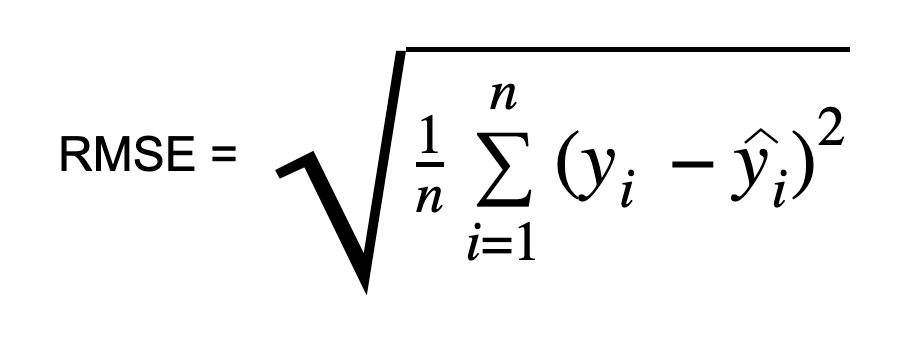
def root_mean_squared_error(true, pred):
squared_error = np.square(true - pred)
sum_squared_error = np.sum(squared_error)
rmse_loss = np.sqrt(sum_squared_error / true.size)
return rmse_loss
Pros of the RMSE Evaluation Metric
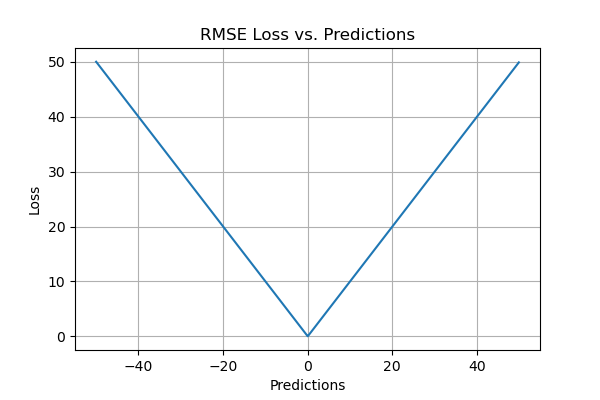
- RMSE is easy to understand.
- It serves as a heuristic for training models.
- It is computationally simple and easily differentiable which many optimization algorithms desire.
- RMSE does not penalize the errors as much as MSE does due to the square root.
Cons of the RMSE Metric
- Like MSE, RMSE is dependent on the scale of the data. It increases in magnitude if the scale of the error increases.
- One major drawback of RMSE is its sensitivity to outliers and the outliers have to be removed for it to function properly.
- RMSE increases with an increase in the size of the test sample. This is an issue when we calculate the results on different test samples.
Relative Squared Error (RSE)
To calculate Relative Squared Error, you take the Mean Squared Error (MSE) and divide it by the square of the difference between the actual and the mean of the data. In other words, we divide the MSE of our model by the MSE of a model that uses the mean as the predicted value.
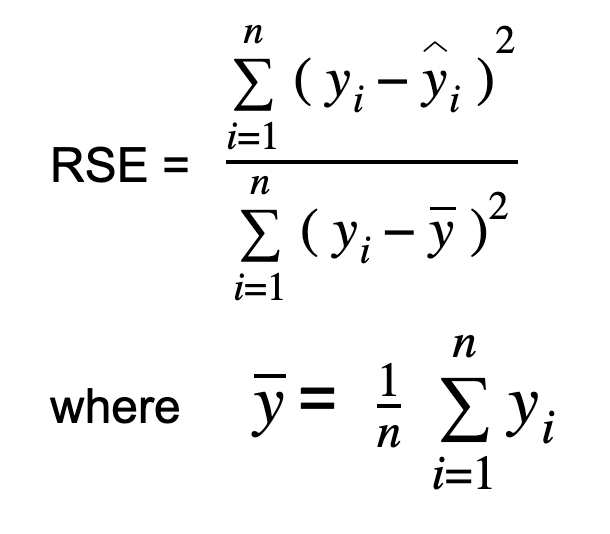
def relative_squared_error(true, pred):
true_mean = np.mean(true)
squared_error_num = np.sum(np.square(true - pred))
squared_error_den = np.sum(np.square(true - true_mean))
rse_loss = squared_error_num / squared_error_den
return rse_loss
The output value of RSE is expressed in terms of ratio. It can range from zero to one. A good model should have a value close to zero while a model with a value greater than 1 is not reasonable.
Pros of the RSE Evaluation Metric
- RSE is not scale-dependent. Hence it can be used to compare models where errors are measured in different units.
- RSE is not sensitive to the mean and the scale of predictions.
Cons of the RSE Evaluation Metric
- RSE does not distinguish between underestimation and overestimation errors, as it only considers the squared differences between y_pred and true values. This means that a model that consistently overestimates or underestimates can still have a low RSE value.
- Like the Mean Squared Error (MSE), RSE is also heavily influenced by outliers in the data points. A few extreme errors can significantly increase the RSE value, even if the model performs well on the majority of the data.
- When the RSE value is much greater than 1, it becomes difficult to interpret the degree of poor performance. An RSE of 2 or 10 indicates that the model performs worse than the mean prediction baseline, but the magnitude of the difference is not clear.
- The interpretation of RSE depends on the performance of the mean prediction baseline for the target values. If the mean prediction itself is a poor baseline, the RSE values may not provide a meaningful comparison.
- Although RSE is scale-independent in terms of the target variable’s units, it can still be sensitive to the scale of the target values. If the target variable has a small range, small errors can result in large RSE values, making the metric less informative.
- For regression analysis problems with strictly non-negative target values (e.g., count data or positive values), the mean prediction baseline may not be a meaningful or appropriate baseline for comparison with the independent variables.
- The interpretation of RSE can also depend on the specific test set used for evaluation. If the test set is not representative of the overall data distribution, the RSE values may not accurately reflect the model’s performance.
Normalized Root Mean Squared Error (NRMSE)
The Normalized RMSE is generally computed by dividing a scalar value. It can be in different ways like,
- RMSE / maximum value in the series
- RMSE / mean
- RMSE / difference between the maximum and the minimum values (if mean is zero)
- RMSE / standard deviation
- RMSE / interquartile range
# implementation of NRMSE with standard deviation
def normalized_root_mean_squared_error(true, pred):
squared_error = np.square((true - pred))
sum_squared_error = np.sum(squared_error)
rmse = np.sqrt(sum_squared_error / true.size)
nrmse_loss = rmse/np.std(pred)
return nrmse_loss
Opting for the interquartile range can be the most suitable choice, especially when dealing with outliers. NRMSE proves effective for comparing models with different dependent variables or when modifications like log transformation or standardization occur. This metric addresses scale-dependency issues, facilitating comparisons across models of varying scales or datasets.
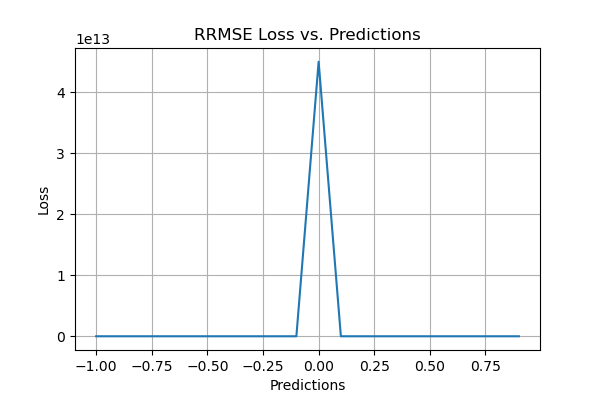
Relative Root Mean Squared Error (RRMSE)
Relative Root Mean Squared Error (RRMSE) is a variant of Root Mean Square Error in Machine Learning (RMSE), gauging predictive model accuracy relative to the target variable range. It normalizes RMSE by the target variable range and presents it as a percentage for easy cross-dataset or cross-variable comparison. RRMSE, a dimensionless form of RMSE, scales residuals against actual values, allowing comparison of different measurement techniques.
- Excellent when RRMSE < 10%
- Good when RRMSE is between 10% and 20%
- Fair when RRMSE is between 20% and 30%
- Poor when RRMSE > 30%
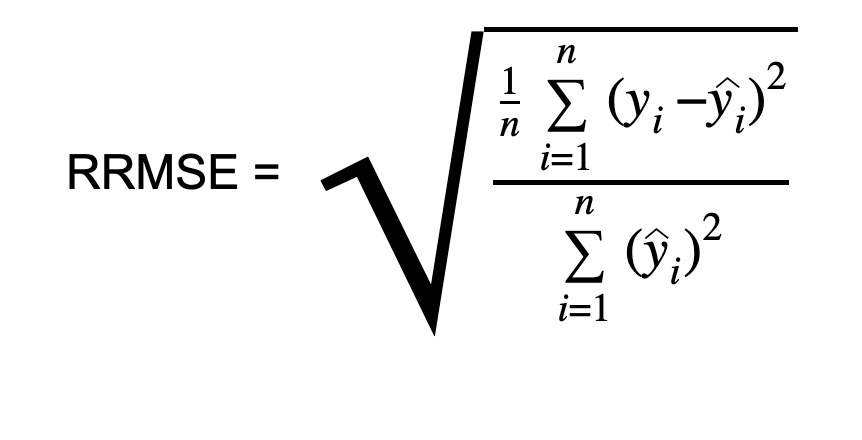
def relative_root_mean_squared_error(true, pred):
num = np.sum(np.square(true - pred))
den = np.sum(np.square(pred))
squared_error = num/den
rrmse_loss = np.sqrt(squared_error)
return rrmse_loss
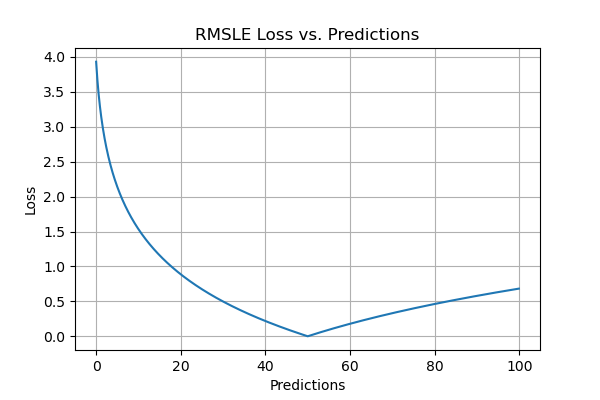
Root Mean Squared Logarithmic Error (RMSLE)
Root Mean Squared Logarithmic Error is calculated by applying log to the actual and the predicted values and then taking their differences. RMSLE is robust to outliers where the small and the large errors are treated evenly.
It penalizes the model more if the predicted value is less than the actual value while the model is less penalized if the predicted value is more than the actual value. It does not penalize high errors due to the log. Hence the model has a larger penalty for underestimation than overestimation. This can be helpful in situations where we are not bothered by overestimation but underestimation is not acceptable.
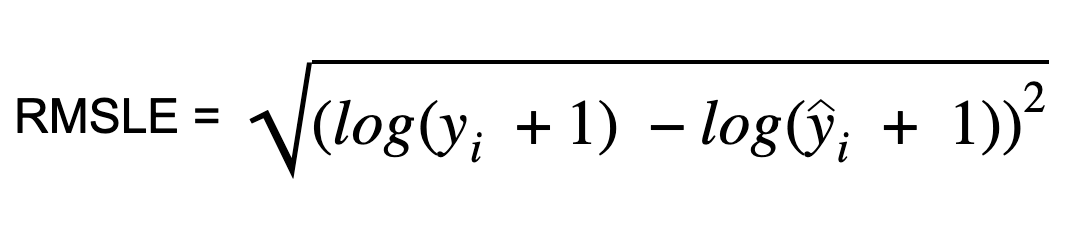
def root_mean_squared_log_error(true, pred):
square_error = np.square((np.log(true + 1) - np.log(pred + 1)))
mean_square_log_error = np.mean(square_error)
rmsle_loss = np.sqrt(mean_square_log_error)
return rmsle_loss
Pros of the RMSLE Evaluation Metric
- RMSLE is not scale-dependent and is useful across a range of scales.
- It is not affected by large outliers.
- It considers only the relative error between the actual value and the predicted value.
Cons of the RMSLE Evaluation Metric
- It has a biased penalty where it penalizes underestimation more than overestimation.
Huber Loss
What if you want a function that learns about the outliers as well as ignores them? Well, Huber loss is the one for you. Huber loss is a combination of both linear and quadratic scoring methods. It has a hyperparameter delta (𝛿) which can be tuned according to the data. The loss will be linear (L1 loss) for values above delta and quadratic (L2 loss) for values below it. It balances and combines good properties of both MAE (Mean Absolute Error) and MSE (Mean Squared Error).
In other words, for loss values less than delta, MSE will be used and for loss values greater than delta, MAE will be used. The choice of delta (𝛿) is extremely critical because it defines our choice of the outlier. Huber loss reduces the weight we put on outliers for larger loss values by using MAE while for smaller loss values it maintains a quadratic function using MSE.
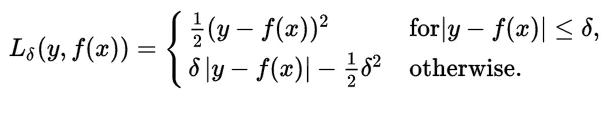
def huber_loss(true, pred, delta):
huber_mse = 0.5 * np.square(true - pred)
huber_mae = delta * (np.abs(true - pred) - 0.5 * (np.square(delta)))
return np.where(np.abs(true - pred) <= delta, huber_mse, huber_mae)
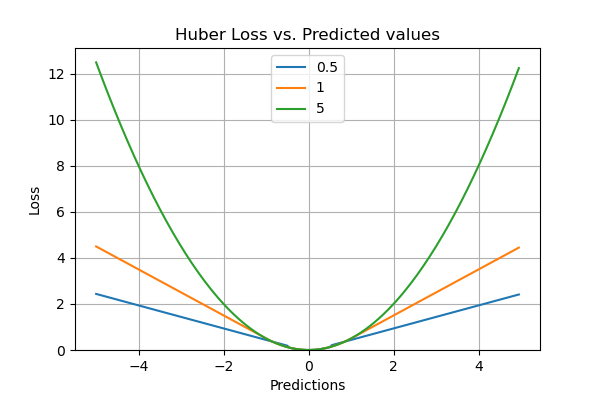
Pros of the Huber Loss Evaluation Metric
- It is differentiable at zero.
- Outliers are handled properly due to the linearity above the delta.
- The hyperparameter, 𝛿 can be tuned to maximize model accuracy.
Cons of the Huber Loss Evaluation Metric
- The additional conditionals and comparisons make Huber loss computationally expensive for large datasets.
- To maximize model accuracy, 𝛿 needs to be optimized and it is an iterative process.
- It is differentiable only once.
Log Cosh Loss
Log cosh calculates the logarithm of the hyperbolic cosine of the error. This function is smoother than quadratic loss. It works like MSE but is not affected by large prediction errors. It is quite similar to Huber loss in the sense that it is a combination of both linear and quadratic scoring methods.
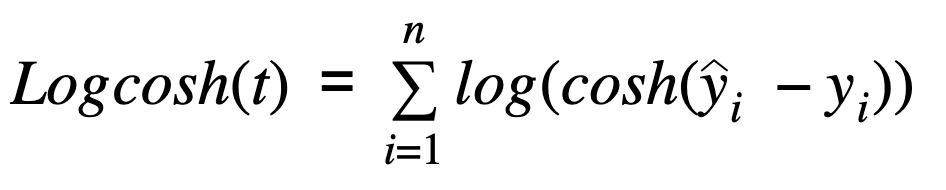
def log_cosh(true, pred):
logcosh = np.log(np.cosh(pred - true))
logcosh_loss = np.sum(logcosh)
return logcosh_loss
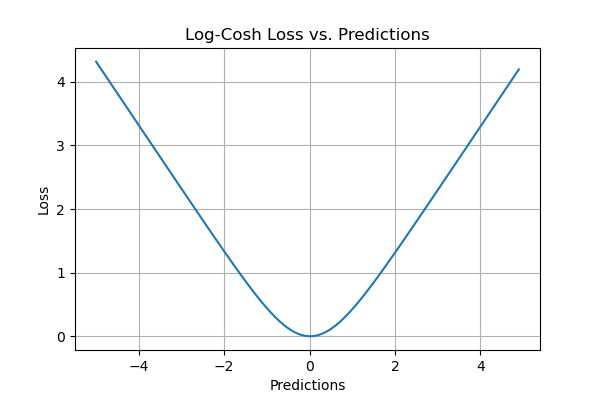
Pros of the Log Cosh Loss Evaluation Metric
- It has the advantages of Huber loss while being twice differentiable everywhere. Some optimization algorithms like XGBoost favor double differentials over functions like Huber which can be differentiable only once.
- It requires fewer computations than Huber.
Cons of the Log Cosh Loss Evaluation Metric
- It is less adaptive as it follows a fixed scale.
- Compared to Huber loss, the derivation is more complex and requires much in-depth study.
Quantile Loss
The quantile regression loss function is applied to predict quantiles. The quantile is the value that determines how many values in the group fall below or above a certain limit. It estimates the conditional median or quantile of the response (dependent) variables across values of the predictor (independent) variables. The loss function is an extension of MAE except for the 50th percentile, where it is MAE. It provides prediction intervals even for residuals with non-constant variance and it does not assume a particular parametric distribution for the response.
γ represents the required quantile. The quantile values are selected based on how we want to weigh the positive and the negative errors. Unlike the squared difference loss used in linear regression models, this loss function is based on absolute differences.
Loss Function
In the loss function above, γ has a value between 0 and 1. When there is an underestimation, the first part of the formula will dominate and for overestimation, the second part will dominate. The chosen value of quantile(γ) gives different penalties for over-prediction and under prediction. When γ = 0.5, underestimation and overestimation are penalized by the same factor, and the median is obtained. When the value of γ is larger, overestimation is penalized more than underestimation. For example, when γ = 0.75 the model will penalize overestimation and it will cost three times as much as underestimation. Optimization algorithms based on gradient descent learn from the quantiles instead of the mean.
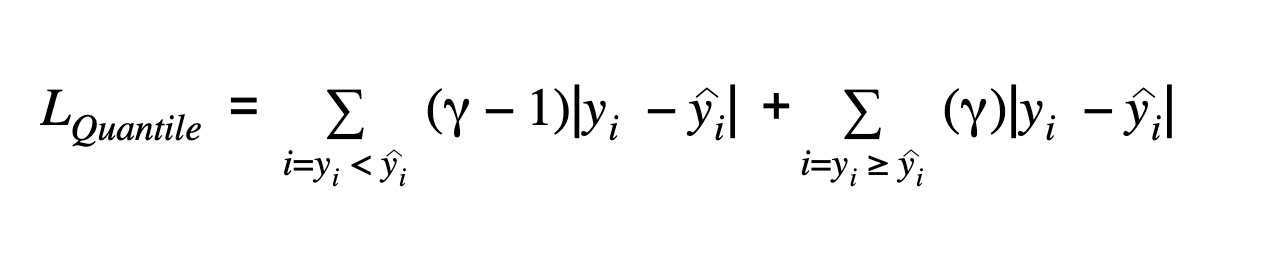
𝛾 represents the required quantile. The quantiles values are selected based on how we want to weigh the positive and the negative errors.
In the loss function above, 𝛾 has a value between 0 and 1. When there is an underestimation, the first part of the formula will dominate and for overestimation, the second part will dominate. The chosen value of quantile(𝛾) gives different penalties for over-prediction and under prediction. When 𝛾 = 0.5, underestimation and overestimation are penalized by the same factor and the median is obtained. When the value of 𝛾 is larger, overestimation is penalized more than underestimation. For example, when 𝛾 = 0.75 the model will penalize overestimation and it will cost three times as much as underestimation. Optimization algorithms based on gradient descent learn from the quantiles instead of the mean.
def quantile_loss(true, pred, gamma):
val1 = gamma * np.abs(true - pred)
val2 = (1-gamma) * np.abs(true - pred)
q_loss = np.where(true >= pred, val1, val2)
return q_loss
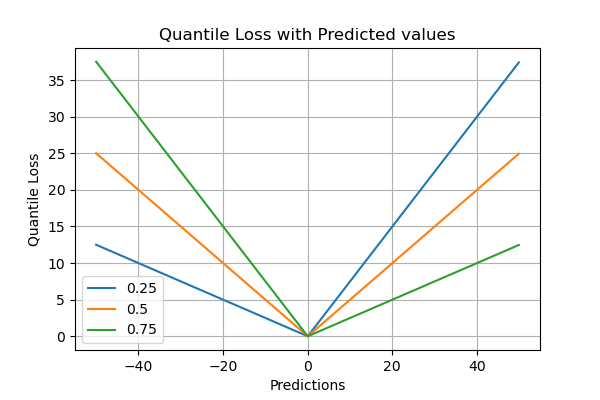
Pros of the Quantile Loss Evaluation Metric
- It is particularly useful when we are predicting an interval instead of point estimates.
- This function can also be used to calculate prediction intervals in neural nets and tree-based models.
- It is robust to outliers.
Cons of the Quantile Loss Evaluation Metric
- Quantile loss is computationally intensive.
- If we use a squared loss to measure the efficiency or if we are to estimate the mean, then quantile loss will be worse.
Conclusion
This comprehensive guide navigated through diverse regression loss functions, shedding light on their applications, advantages, and drawbacks. The article demystified complex metrics like MAE, MBE, RAE, MAPE, MSE, RMSE (the root mean squared error), RSE, NRMSE, RRMSE, and RMSLE, and introduced specialized losses like Huber, Log Cosh, and Quantile. It emphasized the nuanced factors influencing loss function selection, from algorithm types to outlier handling. Additionally, it covered the coefficient of determination (R-squared), and r2_score function from sklearn.metrics import, and adjusted r-squared, which are important evaluation metrics for assessing the performance of machine learning algorithms in regression problems.
Thank you for reading down here! I hope this article was helpful in your learning journey. I would love to hear in the comments about any other loss functions that I have missed. Happy Evaluating!
Key Takeaways
- Loss functions are essential for comparing predicted values with actual values to refine regression models.
- Mean Absolute Error (MAE) is simple to calculate and handles outliers well but is not differentiable at zero.
- Mean Squared Error (MSE) is sensitive to outliers and penalizes larger errors more due to squaring.
- Root Mean Squared Error (RMSE) provides an intuitive measure of model accuracy and is easy to interpret.
- Advanced metrics like Huber Loss and Log Cosh Loss combine properties of MAE and MSE for robust outlier handling.
Frequently Asked Questions
A. The four evaluation metrics are accuracy, precision, recall, and F1 score.
A. Evaluation metrics and scores measure a model’s performance, quantifying its ability to make correct predictions.
A. Evaluation metrics are important because they provide insights into a model’s effectiveness, guiding improvements and ensuring reliable results.
A. In NLP, evaluation metrics assess models based on metrics like BLEU, ROUGE, perplexity, and word error rate, reflecting their linguistic accuracy and coherence.
References
- https://www.sciencedirect.com/topics/engineering/mean-bias-error
- https://www.sciencedirect.com/science/article/abs/pii/S1364032115013258
- https://journals.plos.org/plosntds/article?id=10.1371/journal.pntd.0005295
- https://support.sas.com/resources/papers/proceedings17/SAS0525-2017.pdf
- https://stats.stackexchange.com/questions/39002/when-is-quantile-regression-worse-than-ols








This is an excellent article. I feel it was very well laid out, structured, and easy to understand. One questions on the Python formula for Relative Root Mean Squared Error (RRMSE), is it missing the division by n?
I read through to the end and it was very educative. Thank you. Question: If you were to choose an evaluator for the comparison of predictions from multiple linear and nonlinear models trained using the same data with small outliers, which top three evaluators will you choose and why?
Thanks for the useful resource. Just want to let you know that you missed 1/n part in the Root Mean Squared Logarithmic Error (RMSLE).
In the definition of RRMSE, if the y_i = actual value, are all zero, the RRMSE is square root of 1/n, not 1 Why is the denominator summed, but the numerator averaged? Also, should the denominator include y_i, the actual value, instead of y_hat_i , predicted value?
Shouldn't the denominator in th. relative and normalized rmse be a function of the true values and not predicted values? It looks like the expressions and the python code examples all make the denominators a function of the predicted values.
Thank you. Excellent article clear all doubt