Have you performed RAG over PDFs, Docs, and Reports? Many important documents are not just simple text. Think about research papers, financial reports, or product manuals. They often contain a mix of paragraphs, tables, and other structured elements. This creates a significant challenge for standard Retrieval-Augmented Generation (RAG) systems. Effective RAG on semi-structured data requires more than just basic text splitting. This guide offers a hands-on solution using intelligent unstructured data parsing and an advanced RAG technique known as the multi-vector retriever, all within the LangChain RAG framework.
Table of contents
Need for RAG on Semi-Structured Data
Traditional RAG pipelines often stumble with these mixed-content documents. First, a simple text splitter might chop a table in half, destroying the valuable data within. Second, embedding the raw text of a large table can create noisy, ineffective vectors for semantic search. The language model might never see the right context to answer a user’s question.
We will build a smarter system that intelligently separates text from tables and uses different strategies for storing and retrieving each. This approach ensures our language model gets the precise, complete information it needs to provide accurate answers.
The Solution: A Smarter Approach to Retrieval
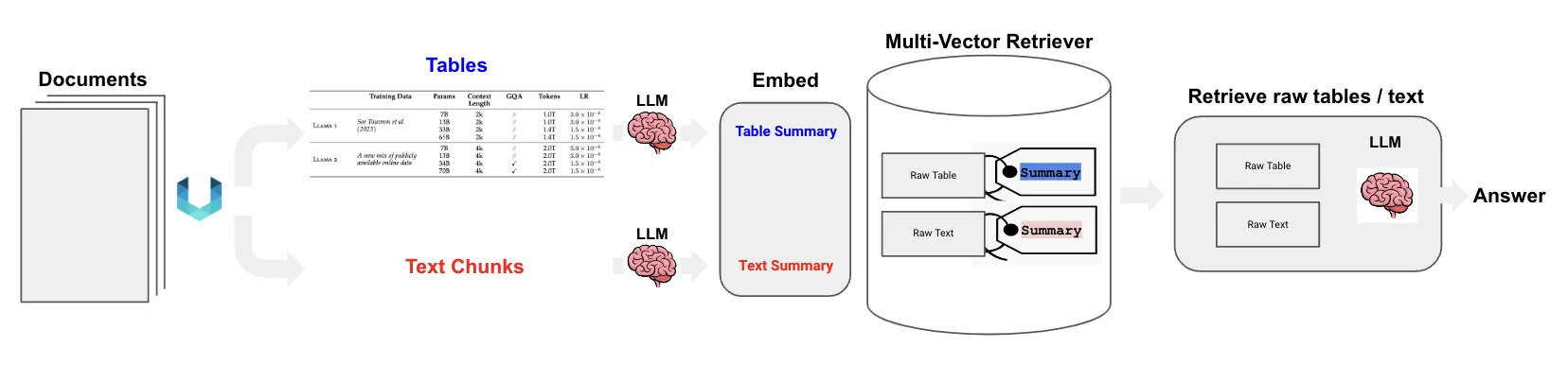
Our solution tackles the core challenges head-on by using two key components. This method is all about preparing and retrieving data in a way that preserves its original meaning and structure.
- Intelligent Data Parsing: We use the Unstructured library to do the initial heavy lifting. Instead of blindly splitting text, Unstructured’s
partition_pdffunction analyzes a document’s layout. It can tell the difference between a paragraph and a table, extracting each element cleanly and preserving its integrity. - The Multi-Vector Retriever: This is the core of our advanced RAG technique. The multi-vector retriever allows us to store multiple representations of our data. For retrieval, we will use concise summaries of our text chunks and tables. These smaller summaries are much better for embedding and similarity search. For answer generation, we will pass the full, raw table or text chunk to the language model. This gives the model the complete context it needs.
The overall workflow looks like this:
Building the RAG Pipeline
Let’s walk through how to build this system step-by-step. We will use the LLaMA2 research paper as our example document.
Step 1: Setting Up the Environment
First, we need to install the necessary Python packages. We’ll use LangChain for the core framework, Unstructured for parsing, and Chroma for our vector store.
! pip install langchain langchain-chroma "unstructured[all-docs]" pydantic lxml langchainhub langchain_openai -qUnstructured’s PDF parsing relies on a couple of external tools for processing and Optical Character Recognition (OCR). If you’re on a Mac, you can install them easily using Homebrew.
!apt-get install -y tesseract-ocr
!apt-get install -y poppler-utilsStep 2: Data Loading and Parsing with Unstructured
Our first task is to process the PDF. We use partition_pdf from Unstructured, which is purpose-built for this kind of unstructured data parsing. We will configure it to identify tables and chunk the document’s text by its titles and subtitles.
from typing import Any
from pydantic import BaseModel
from unstructured.partition.pdf import partition_pdf
# Get elements
raw_pdf_elements = partition_pdf(
filename="/content/LLaMA2.pdf",
# Unstructured first finds embedded image blocks
extract_images_in_pdf=False,
# Use layout model (YOLOX) to get bounding boxes (for tables) and find titles
# Titles are any sub-section of the document
infer_table_structure=True,
# Post processing to aggregate text once we have the title
chunking_strategy="by_title",
# Chunking params to aggregate text blocks
# Attempt to create a new chunk 3800 chars
# Attempt to keep chunks > 2000 chars
max_characters=4000,
new_after_n_chars=3800,
combine_text_under_n_chars=2000,
image_output_dir_path=path,
)After running the partitioner, we can see what types of elements it found. The output shows two main types: CompositeElement for our text chunks and Table for the tables.
# Create a dictionary to store counts of each type
category_counts = {}
for element in raw_pdf_elements:
category = str(type(element))
if category in category_counts:
category_counts[category] += 1
else:
category_counts[category] = 1
# Unique_categories will have unique elements
unique_categories = set(category_counts.keys())
category_countsOutput:

As you can see, Unstructured did a great job identifying 2 distinct tables and 85 text chunks. Now, let’s separate these into distinct lists for easier processing.
class Element(BaseModel):
type: str
text: Any
# Categorize by type
categorized_elements = []
for element in raw_pdf_elements:
if "unstructured.documents.elements.Table" in str(type(element)):
categorized_elements.append(Element(type="table", text=str(element)))
elif "unstructured.documents.elements.CompositeElement" in str(type(element)):
categorized_elements.append(Element(type="text", text=str(element)))
# Tables
table_elements = [e for e in categorized_elements if e.type == "table"]
print(len(table_elements))
# Text
text_elements = [e for e in categorized_elements if e.type == "text"]
print(len(text_elements))Output:

Step 3: Creating Summaries for Better Retrieval
Large tables and long text blocks don’t create very effective embeddings for semantic search. A concise summary, however, is perfect. This is the central idea of using a multi-vector retriever. We’ll create a simple LangChain chain to generate these summaries.
from langchain_core.output_parsers import StrOutputParser
from langchain_core.prompts import ChatPromptTemplate
from langchain_openai import ChatOpenAI
from getpass import getpass
OPENAI_KEY = getpass('Enter Open AI API Key: ')
LANGCHAIN_API_KEY = getpass('Enter Langchain API Key: ')
LANGCHAIN_TRACING_V2="true"
# Prompt
prompt_text = """You are an assistant tasked with summarizing tables and text. Give a concise summary of the table or text. Table or text chunk: {element} """
prompt = ChatPromptTemplate.from_template(prompt_text)
# Summary chain
model = ChatOpenAI(temperature=0, model="gpt-4.1-mini")
summarize_chain = {"element": lambda x: x} | prompt | model | StrOutputParser()Now, we apply this chain to our extracted tables and text chunks. The batch method allows us to process these concurrently, which speeds things up.
# Apply to tables
tables = [i.text for i in table_elements]
table_summaries = summarize_chain.batch(tables, {"max_concurrency": 5})
# Apply to texts
texts = [i.text for i in text_elements]
text_summaries = summarize_chain.batch(texts, {"max_concurrency": 5})Step 4: Building the Multi-Vector Retriever
With our summaries ready, it’s time to build the retriever. It uses two storage components:
- A vectorstore (ChromaDB) stores the embedded summaries.
- A docstore (a simple in-memory store) holds the raw table and text content.
The retriever uses unique IDs to create a link between a summary in the vector store and its corresponding raw document in the docstore.
import uuid
from langchain.retrievers.multi_vector import MultiVectorRetriever
from langchain.storage import InMemoryStore
from langchain_chroma import Chroma
from langchain_core.documents import Document
from langchain_openai import OpenAIEmbeddings
# The vectorstore to use to index the child chunks
vectorstore = Chroma(collection_name="summaries", embedding_function=OpenAIEmbeddings())
# The storage layer for the parent documents
store = InMemoryStore()
id_key = "doc_id"
# The retriever (empty to start)
retriever = MultiVectorRetriever(
vectorstore=vectorstore,
docstore=store,
id_key=id_key,
)
# Add texts
doc_ids = [str(uuid.uuid4()) for _ in texts]
summary_texts = [
Document(page_content=s, metadata={id_key: doc_ids[i]})
for i, s in enumerate(text_summaries)
]
retriever.vectorstore.add_documents(summary_texts)
retriever.docstore.mset(list(zip(doc_ids, texts)))
# Add tables
table_ids = [str(uuid.uuid4()) for _ in tables]
summary_tables = [
Document(page_content=s, metadata={id_key: table_ids[i]})
for i, s in enumerate(table_summaries)
]
retriever.vectorstore.add_documents(summary_tables)
retriever.docstore.mset(list(zip(table_ids, tables)))Step 5: Running the RAG Chain
Finally, we construct the complete LangChain RAG pipeline. The chain will take a question, use our retriever to fetch the relevant summaries, pull the corresponding raw documents, and then pass everything to the language model to generate an answer.
from langchain_core.runnables import RunnablePassthrough
# Prompt template
template = """Answer the question based only on the following context, which can include text and tables:
{context}
Question: {question}
"""
prompt = ChatPromptTemplate.from_template(template)
# LLM
model = ChatOpenAI(temperature=0, model="gpt-4")
# RAG pipeline
chain = (
{"context": retriever, "question": RunnablePassthrough()}
| prompt
| model
| StrOutputParser()
)
Let's test it with a specific question that can only be answered by looking at a table in the paper.
chain.invoke("What is the number of training tokens for LLaMA2?")Output:

The system works perfectly. By inspecting the process, we can see that the retriever first found the summary of Table 1, which discusses model parameters and training data. Then, it retrieved the full, raw table from the docstore and provided it to the LLM. This gave the model the exact data needed to answer the question correctly, proving the power of this RAG on semi-structured data approach.
You can access the full code on the Colab notebook or the GitHub repository.
Conclusion
Handling documents with mixed text and tables is a common, real-world problem. A simple RAG pipeline is not enough in most cases. By combining intelligent unstructured data parsing with the multi-vector retriever, we create a much more robust and accurate system. This method ensures that the complex structure of your documents becomes a strength, not a weakness. It provides the language model with complete context in an easy-to-understand manner, leading to better, more reliable answers.
Read more: Build a RAG Pipeline using Llama Index
Frequently Asked Questions
A. Yes, the Unstructured library supports a wide range of file types. You can simply swap the partition_pdf function with the appropriate one, like partition_docx.
A. No, you could generate hypothetical questions from each chunk or simply embed the raw text if it’s small enough. A summary is often the most effective for complex tables.
A. Large tables can create “noisy” embeddings where the core meaning is lost in the details. This makes semantic search less effective. A concise summary captures the essence of the table for better retrieval.





